J5: Micrograph Denoiser (Prerun)
Inputs
No inputs
Custom Parameters
All default parameters
J6: Curate Exposures
Inputs
| J5 | exposures |
|---|
Custom Parameters
All default parameters
J7: Blob Picker
Inputs
| J6 | exposures accepted |
|---|
Custom Parameters
| Minimum Search Diameter (A) | 200 |
|---|---|
| Maximum Search Diameter (A) | 320 |
| Use denoised micrographs | |
| Min. separation dist (diameters) | 0.5 |
J8: Junk Detector
Custom Parameters
All default parameters
J9: Inspect Picks
Custom Parameters
| Calibrate Pick score to CTF | |
|---|---|
| Calibrate Power score to CTF | |
| Auto Cluster | |
| Target power score | 200 |
J10: Extract from Micrographs (Blob Picks)
Custom Parameters
| Number of CPU cores | 8 |
|---|---|
| Extraction box size (pix) | 450 |
| Fourier crop to box size (pix) | 64 |
| Save results in 16-bit floating point |
J11: 2D Classification (Generate Templates)
Inputs
| J10 | particles |
|---|
Custom Parameters
| Number of 2D classes | 40 |
|---|---|
| Initial classification uncertainty factor | 0.5 |
| Maximum resolution (A) | 15 |
| 2D zeropad factor | 1 |
J12: Select 2D (Template Images)
Custom Parameters
All default parameters
J13: Template Picker
Custom Parameters
| Use CTFs to filter the templates | |
|---|---|
| Use denoised micrographs | |
| Minimum Search Diameter (A) | 300 |
| Min. separation dist (diameters) | 0.6 |
J14: Junk Detector
Custom Parameters
All default parameters
J15: Inspect Picks
Custom Parameters
| Calibrate Pick score to CTF | |
|---|---|
| Calibrate Power score to CTF |
J16: Extract from Micrographs (Template Picks)
Custom Parameters
| Number of CPU cores | 8 |
|---|---|
| Extraction box size (pix) | 450 |
| Fourier crop to box size (pix) | 64 |
| Save results in 16-bit floating point |
J17: 2D Classification (pre-3D)
Inputs
| J16 | particles |
|---|
Custom Parameters
| Number of 2D classes | 80 |
|---|---|
| Maximum resolution (A) | 15 |
| Initial classification uncertainty factor | 0.5 |
| 2D zeropad factor | 1 |
J18: Select 2D (pre-3D)
Custom Parameters
All default parameters
J19: Ab Initio Reconstruction (bad volumes)
Inputs
| J18 | particles excluded |
|---|
Custom Parameters
| Number of Ab-Initio classes | 3 |
|---|---|
| Number of initial iterations | 20 |
| Number of final iterations | 30 |
J20: Ab Initio Reconstruction (good volume)
Inputs
| J18 | particles selected |
|---|
Custom Parameters
All default parameters
J21: Heterogeneous Refinement
Custom Parameters
| Refinement box size (Voxels) | 64 |
|---|
J22: Extract from Micrographs (3D Curated)
Custom Parameters
| Extraction box size (pix) | 450 |
|---|---|
| Number of CPU cores | 8 |
| Fourier crop to box size (pix) | 300 |
| Save results in 16-bit floating point |
J23: Volume Tools (flip hand)
Inputs
| J21 | volume class 0 |
|---|
Custom Parameters
| Flip hand |
|---|
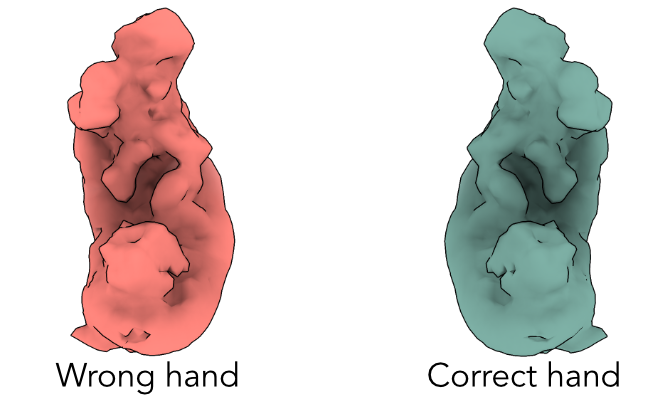
J24: Non-Uniform Refinement (consensus)
Custom Parameters
| Minimize over per-particle scale | |
|---|---|
| Dynamic mask start resolution (A) | 1 |
J25: Volume Tools (lowpass)
Inputs
| J24 | volume |
|---|
Custom Parameters
| Lowpass filter (A) | 15 |
|---|
J26: Volume Tools (threshold)
Inputs
| J25 | volume |
|---|
Custom Parameters
| Type of output volume | mask |
|---|---|
| Threshold (must set to dilate, pad, fill holes, or z-clip) | {dataset dependent} |
| Dilation radius (pix) | 4 |
J27: 3D Variability Analysis
Custom Parameters
| Number of modes to solve | 1 |
|---|---|
| Filter resolution (A) | 12 |
J28: 3D Variability Display (simple)
Custom Parameters
| Downsample to box size | 128 |
|---|---|
| Crop to size (after downsample) | 100 |
J29: 3D Variability Display (cluster)
Custom Parameters
| Downsample to box size | 128 |
|---|---|
| Crop to size (after downsample) | 100 |
| Output mode | cluster |
| Number of frames/clusters | 2 |
Opening 3DVA volume series
- Download both jobs' "All volumes" series outputs.
- Unzip them
- Rename the directories to "simple" and "cluster"
- Run the commands below
open ~/Downloads/simple/*.mrc vseries true
open ~/Downloads/cluster/*.mrc vseries true
J30: 3D Classification
Custom Parameters
| Number of classes | 2 |
|---|---|
| Filter resolution (Å) | 10 |
J31: Non-Uniform Refinement (class 0)
Custom Parameters
| Minimize over per-particle scale | |
|---|---|
| Dynamic mask start resolution (A) | 1 |
J32: Non-Uniform Refinement (class 1)
Custom Parameters
| Minimize over per-particle scale | |
|---|---|
| Dynamic mask start resolution (A) | 1 |
Comparing with an atomic model
-
Open the active state model by running
open 8OLT -
Use the "translate selected models" right mouse mode, either by
clicking it in the GUI or running
ui mousemode right 'translate selected models' - Select the model and move it into the appropriate position
-
Fit the model into the active map using
fitmap #{modelID} inMap #{activeID}
where each ID is replaced with the appropriate number